Protein conformational dynamics studied by 15N and 1H R1ρ relaxation dispersion: Application to wild-type and G53A ubiquitin crystals
14-Apr-2017
Solid State Nuclear Magnetic Resonance, Volume 87, Pages 86-95, https://doi.org/10.1016/j.ssnmr.2017.04.002
Solid State Nuclear Magnetic Resonance, online article
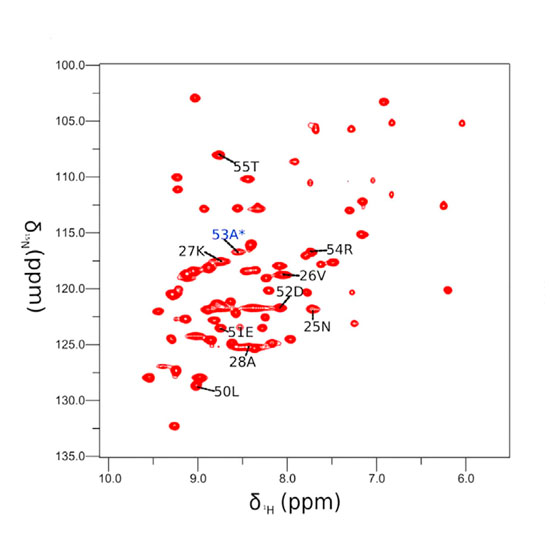
Solid-state NMR spectroscopy can provide site-resolved information about protein dynamics over many time scales. Here we combine protein deuteration, fast magic-angle spinning (~45–60 kHz) and proton detection to study dynamics of ubiquitin in microcrystals, and in particular a mutant in a region that undergoes microsecond motions in a β-turn region in the wild-type protein. We use 15N R1ρ relaxation measurements as a function of the radio-frequency (RF) field strength, i.e. relaxation dispersion, to probe how the G53A mutation alters these dynamics. We report a population-inversion of conformational states: the conformation that in the wild-type protein is populated only sparsely becomes the predominant state. We furthermore explore the potential to use amide-1H R1ρ relaxation to obtain insight into dynamics. We show that while quantitative interpretation of 1H relaxation remains beyond reach under the experimental conditions, due to coherent contributions to decay, one may extract qualitative information about flexibility.